
Since the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) originally emerged in December 2019, numerous global efforts have been made to develop therapeutic drugs to either reduce the severity of symptoms associated with the coronavirus disease 2019 (COVID-19). In a recent study under consideration at the journal Critical Care and currently posted to the Research Square* preprint server, researchers identify a methodology and workflow to repurpose drugs against the cyclin-dependent kinase 6 (CDK6) target gene of SARS-CoV-2.
Study: High neutrophils and triglycerides lead to critical illness in 2 COVID-19 and reveal CDK6 inhibitors as potential treatment. Image Credit: StudioMolekuul / Shutterstock.com
Background
Despite the efforts that have been made by researchers around the world to advance treatment options for COVID-19, only a select few of these treatment modalities have reached clinical trials. Thus, many researchers have looked towards repurposing existing drugs to accelerate the drug discovery process and make these agents more readily available to the public with reduced costs and time.
Critical illness in COVID-19 represents high lung infiltration with immune cells, macrophages, and neutrophils, leading to respiratory disorders and other pathologies. Thus, understanding the molecular targets and host genetics, as well as their mechanism in uncontrolled immune reactions to SARS-CoV-2 infection, can assist in selecting appropriate drugs to treat COVID-19.
About the study
The current study included data on COVID-19-positive patients obtained from the United Kingdom Biobank project. The infectious disease phenotype was developed for respiratory infections, acute respiratory distress syndrome (ARDS), influenza, and pneumonia with hospitalization or death, which included a total of 42,065 patients.
The workflow was designed to identify traits that lead to critical illness in patients. The U.K. Biobank data was divided into one cohort with critically ill patients and a second cohort with mild or no symptoms, which amounted to a total of 1,505 cases and controls.
Patients who were hospitalized/ventilated or died of COVID-19 were considered critically ill, whereas those who were ill but survived after the infection were defined as controls. Age and sex were also taken into consideration while screening.
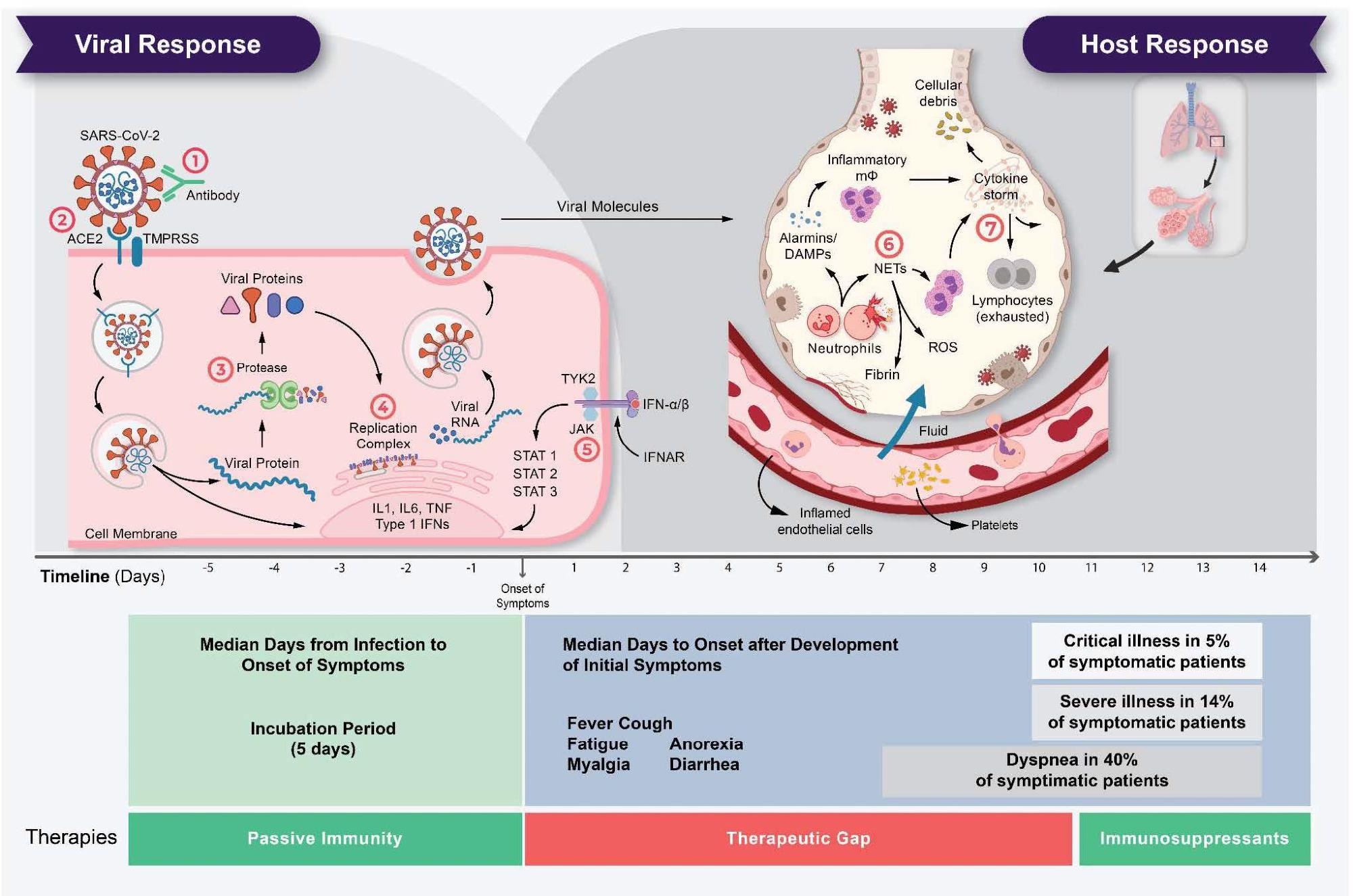 The life cycle of SARS-CoV-2 and the corresponding pathogenesis of COVID- 19 display two phases: a viral response and a host-response phase. In the viral response phase, the virus enters the host cell and viral replication begins. Approximately five days after infection and successful replication, initial mild and moderate symptoms such as fever, cough, fatigue, anorexia, myalgia, and diarrhea are observed in conjunction with a decrease in lymphocyte cell count (lymphopenia). The following host-response phase determines the severity of the disease: in some patients, uncontrolled overreaction of the immune system – so-called virus-induced immunopathology – requires hospitalization and respiratory support due to acute respiratory distress syndrome (ARDS). Thus, severe cases of COVID-19 originate from an immune overreaction rather than from the viral infection itself. Currently, there are seven drug mechanisms described: ! Passive immunity; ” Entry inhibitors; # Protease inhibitors; $ Polymerase inhibitors; % JAK inhibitors; & NETosis inhibitors; ‘ Immunosuppressants.
The life cycle of SARS-CoV-2 and the corresponding pathogenesis of COVID- 19 display two phases: a viral response and a host-response phase. In the viral response phase, the virus enters the host cell and viral replication begins. Approximately five days after infection and successful replication, initial mild and moderate symptoms such as fever, cough, fatigue, anorexia, myalgia, and diarrhea are observed in conjunction with a decrease in lymphocyte cell count (lymphopenia). The following host-response phase determines the severity of the disease: in some patients, uncontrolled overreaction of the immune system – so-called virus-induced immunopathology – requires hospitalization and respiratory support due to acute respiratory distress syndrome (ARDS). Thus, severe cases of COVID-19 originate from an immune overreaction rather than from the viral infection itself. Currently, there are seven drug mechanisms described: ! Passive immunity; ” Entry inhibitors; # Protease inhibitors; $ Polymerase inhibitors; % JAK inhibitors; & NETosis inhibitors; ‘ Immunosuppressants.
Study findings
The difference between the infectious disease and healthy cohorts was determined by identifying 64 candidate predictive traits from the U.K. Biobank biological sample data. Screened traits included 33 blood cells, 89 types, 30 blood biochemistries, and body mass index (BMI), all of which were determined several years before infection.
The traits were identified by performing t-tests and Mann-Whitney U-test, which showed differences in 53 traits. The researchers also performed regression modeling to determine the effect of these 53 traits on critically ill COVID-19 patients. Taken together, 21 traits were found to significantly affect the severity of illness.
The treatment effect on critical illness was determined by performing propensity score analysis, wherein 11 traits were found to significantly affect COVID-19 severity, irrespective of other independent traits.
Drop 1 analysis showed that out of seven independent traits including BMI, neutrophil cell count, cystatin C, glucose, glycated hemoglobin, triglycerides, and five traits related to reticulocytes, neutrophils cell count and triglycerides contributed significantly to the illness.
Neutrophils and triglycerides identified in the Drop 1 analysis were studied for their genetics using genome-wide association (GWA) analysis. This analysis was used to determine the pathogenesis of the diseases to find the drug targets.
Triglycerides regulate neutrophil cell levels, which respond to the CDK6 gene that encodes for CDK6. Thus, inhibiting CDK6 may reduce neutrophil counts, further decreasing the immune hyper-reaction.
Conclusions
In the current study, researchers aimed to determine the relationship between high neutrophil cell counts and triglycerides that may contribute to critical COVID-19. To this end, the researchers designed a workflow that determined the causal relationship between triglycerides and neutrophil count as a predisposition for immune hyper reaction in COVID-19.
The current findings indicate that CDK6 is a potential target against neutrophils and that CDK4/6 inhibitors may have potential in SARS-CoV-2 induced immune pathogenesis. However, to ascertain this possibility, further clinical studies are warranted.
*Important notice
Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.