
Several prophylactic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccines are being used across the globe to control the spread of the COVID-19 pandemic. However, a successful control strategy requires the implementation of effective treatment options as well, which are still lacking against the coronavirus disease 19 (COVID-19).
The use of virus-neutralizing monoclonal antibodies (MAbs) is a hopeful treatment intervention. However, producing safe and effective mAbs remains a complex task, and the duration of lasting protection remains unknown. Simultaneously, mutations in the constantly evolving SARS-CoV-2 may result in an escape from Mab recognition and neutralization. Therefore, the search for a broad-spectrum therapeutic agent against COVID-19 continues.
In their recent study posted to the preprint server bioRxiv*, the researchers from China have shown that tuftsin, a natural phagocytosis-stimulating peptide, can effectively impair the binding of SARS-CoV-2 spike S1 subunit to ACE2 receptor in a dose-dependent manner and therefore can be considered a potential drug against SARS-CoV-2 infection.
 Study: Tuftsin: a natural molecule against SARS-CoV-2 infection. Image Credit: NIAID
Study: Tuftsin: a natural molecule against SARS-CoV-2 infection. Image Credit: NIAID
Tuftsin and its targets
Neuropilin 1 (NRP1), an important receptor of the natural peptide tuftsin, has been found to facilitate SARS-CoV-2 entry into the cells in the presence of ACE2. Tuftsin itself is a natural phagocytosis-stimulating tetrapeptide released from the Fc fragment of the heavy chain of the Immunoglobulin G (IgG). It is immunostimulatory and stimulates phagocytic cells, especially macrophages. Being a stable natural immune stimulating peptide molecule with a low toxicity in vitro and in vivo, tuftsin is already an attractive candidate for immunotherapy.
Based on the fact that SARS-CoV-2 and tuftsin maintain the same receptor, NRP1, the team from China hypothesized that tuftsin could potentially inhibit SARS-CoV-2 infection by competitively interacting with NRP1, and subsequently conducted the current study to verify.
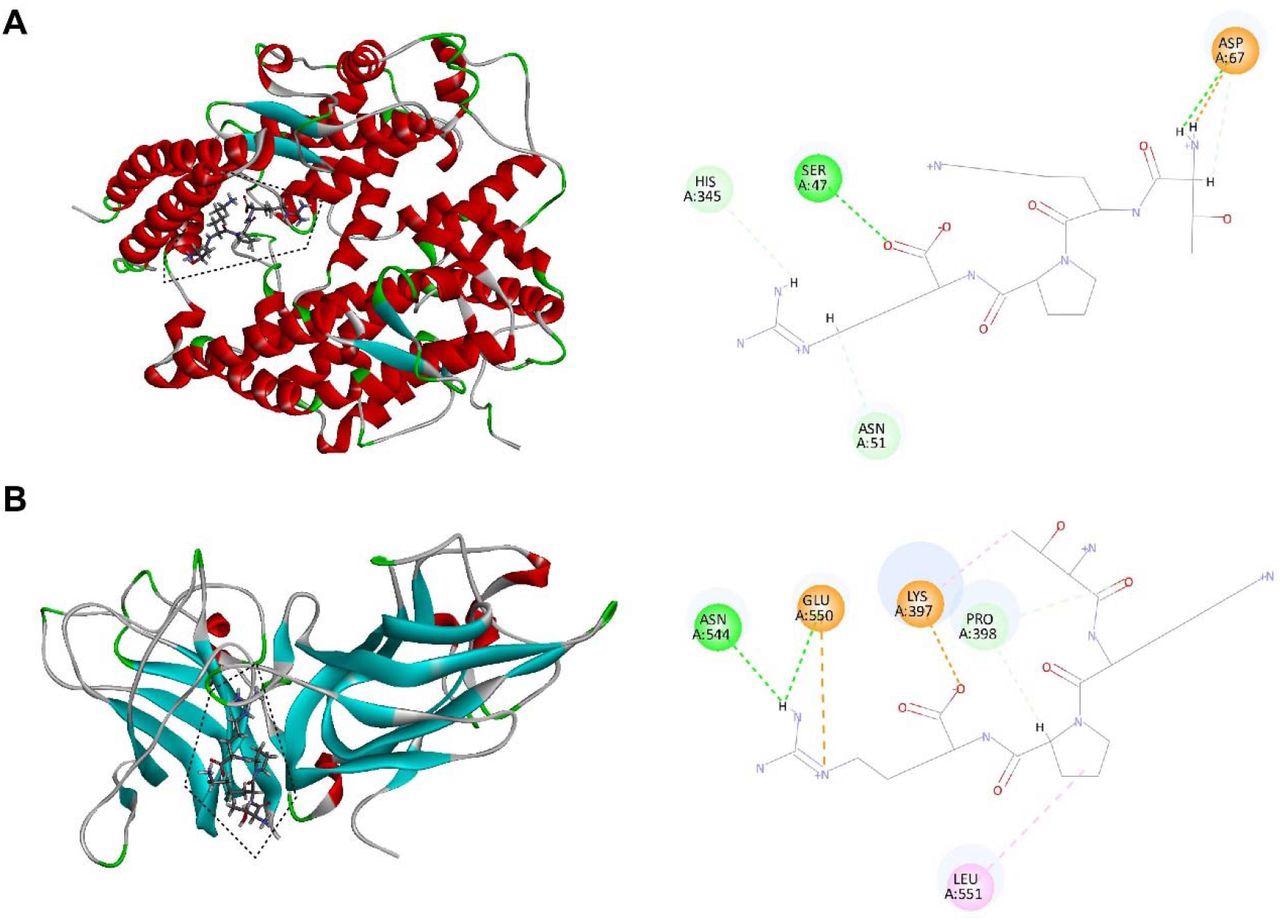
Molecular interaction of tuftsin with ACE2 and NRP1. (A) (Left) The binding pattern of tuftsin with ACE2. Binding area was circled by black dotted line. Secondary structural elements are depicted as ribbons (coils, α-helices, arrows, β-sheets). Color is based on secondary structures (α-helices, red; β-sheets, skyblue; loops, green). (Right) Molecular interaction schemes of tuftsin with the relative residues of ACE2. Green lines represent conventional hydrogen bonds; light green lines represent carbon hydrogen bonds; orange lines represent salt bridges; and pink lines represent alkyl bonds. (B) (Left) The binding pattern of tuftsin with NRP1. Binding area was circled by black dotted line. (Right) Molecular interaction schemes of tuftsin with the relative residues of NRP1. Other interpretations are the same as above.
Bioinformatics analysis and molecular docking show that in addition to NRP1, tuftsin also targets SARS-CoV-2 receptor ACE2
The publicly available 2D structure of tuftsin was retrieved, and its 3D structure was built through molecular simulation. The team then collected the known and potential targets of tuftsin, along with the disease-associated genes of COVID-19 for comparison/ intersection analysis.
Notably, a total of 91 intersecting proteins were found between tuftsin targets and COVID-19-associated genes. Overlapping proteins were mainly found to be associated with immune functions.
Bioinformatic COVID-19 pathway enrichment analysis demonstrated that the SARS-CoV-2 receptors ACE2 and NRP1 were also the targets of tuftsin, highlighting its potential to become an anti-COVID-19 drug that acts via a competitive mechanism.
Later, the team performed molecular docking to determine the potential binding sites and binding affinity between tuftsin and its newly reported target, ACE2. The results showed that the affinity of tuftsin and ACE2 was -6.9 kcal/mol, signifying that they could combine spontaneously. Notably, the binding sites of tuftsin and ACE2 were adjacent to the binding site of S1-RBD and ACE2, indicating that tuftsin could competitively inhibit S1 binding to ACE2 by covering their binding sites.
Similarly, the potential binding sites and binding affinity between tuftsin and its already known receptor, NRP1, were also determined. The docking results showed a high binding affinity of -8.1 kcal/mol between tuftsin and NRP1. In addition, the binding region of tuftsin and NRP1 overlapped with the binding site of NRP1 and S1-RBD.
Collectively, the molecular docking demonstrated that tuftsin could bind ACE2 and NRP1 and inhibit the SARS-CoV-2 S1 binding of ACE2 and NRP1 by covering their interactional sites.
Surface plasmon resonance (SPR) analyses confirm the direct binding of Tuftsin with ACE2 and NPR1
To determine the kinetics of the binding reaction of tuftsin with ACE2 and NPR1, different concentrations of tuftsin were injected over recombinant human ACE2 immobilized on one half of the chip surface and over recombinant human NRP1 on another half of the chip surface. The results showed that tuftsin specifically bound ACE2 and NRP1 with an equilibrium dissociation constant (KD) of 460 μmol/L and 10.65 μmol/L, respectively. Being a gold standard for detecting drug-target interactions, the demonstration of the direct and specific binding of tuftsin with ACE2 and NRP1 by SPR validated the results of bioinformatics analyses and molecular docking assays.
Tuftsin competitively impairs the binding of SARS-CoV-2 S1 to ACE2
The team performed an SPR-based competition assay which confirmed that tuftsin effectively impaired the binding of SARS-CoV-2 S1 to ACE2 in a dose-dependent manner. Tuftsin with a concentration 9 μM had slight inhibitory activity on binding. Increasing the concentration to 156 μM significantly reduced the binding of SARS-CoV-2 S1 and ACE2. Further, when the concentration was increased to 625 μM, the binding of SARS-CoV-2 S1 and ACE2 was blocked entirely.
Implications
Based on the findings, the team advises on using tuftsin as an oral or nasal spray. The tuftsin spray could efficiently protect the upper respiratory tract, which the antibodies induced by vaccines cannot effectively protect.
Notably, people with mild and asymptomatic COVID-19 seem to clear the virus faster due to comparatively more robust innate and adaptive immune responses. The team speculates that tuftsin, being a human natural immunostimulating peptide with significant roles related to innate immunity, and now reported to target the important receptors of SARS-CoV-2 S1, might hold some crucial roles in asymptomatic carriers or mild cases of COVID-19.
*Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.